
Protai, an Israeli company specializing in artificial intelligence-based drug discovery, has announced an $8 million (€7 million) round of funding from venture capital firms Grove Ventures and Pitango HealthTech. The start-up, which combines deep proteomics with machine learning, will be able to accelerate its development, the implementation of research programs and strengthen partnerships with pharmaceutical companies thanks to this round of financing.
Proteomics consists of studying all the proteins in an organism, a biological fluid, a tissue, a cell or even a cell compartment. This set of proteins is called proteome. Its study allows us to understand the molecular mechanisms of essential cellular functions as well as the physiology of living beings. Proteomic analysis is applied to the discovery of new therapeutic targets and to the study of drug effects.
Protai’s proteomic atlas
Protai has completed “the largest and most robust atlas today” in less than a year, with more than 50,000 clinical data samples and healthy samples as a baseline for pattern recognition.
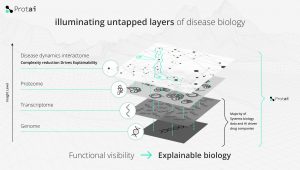
The harmonization of this chaotic data allowed the creation of a map of proteins and their interactions within a given disease and to understand the molecular mechanisms.
Eran Seger, CEO of Protai said:
“Our improvements in biological explicability are resulting in better drug targets and drug candidates. In our lung cancer proof of concept, we have identified several high-value targets that we are currently pursuing in our independent drug program, as well as other markers that will help expand the use of known drugs in this indication.”
By leveraging mass spectrometry proteomics and other large-scale protein-focused datasets, Protai is building machine learning models to identify up to 1,000,000 proteoforms.
Discovering new drugs
Protai thus uses biology, chemistry and machine learning, a multidisciplinary approach, to predict which drug searches to pursue. Artificial intelligence is used to determine the structure and functionality of proteins, but the heterogeneity of proteomes complicates the development of proteomics. The latter and the discovery of new drugs that it could lead to have become crucial issues: the overuse of antibiotics has developed resistance in bacteria to their administration, leading to a global public health problem. Most of the major pharmaceutical companies are therefore opening their own proteomics division or surrounding themselves with experts in this field to find alternatives. All of this research will lead to a better understanding of human biology and improve the medical care of patients (diagnosis, therapeutic choices, etc.).
A sharing of knowledge
Models capable of predicting the structure of proteins have already been developed: AlphaFold by Deepmind (a subsidiary of Alphabet) and RoseTTAFold by the University of Washington with the support of Microsoft, among others. The databases of these models are freely available online.
In France, three proteomics sites, based respectively in Grenoble, Toulouse and Strasbourg, work together within the framework of the national proteomics infrastructure ProFi. They share their experiences, develop common software and protocols to improve the analysis and processing of proteomic data and propose this work to the French and international scientific community.
Translated from Levée de fonds de 7 millions d’euros pour Protai, plateforme de découverte de médicaments basée sur l’IA et centrée sur la protéomique









