In a collaborative research between the German research center Helmholtz Zentrum München (HZM) and Facebook AI, an artificial intelligence model was designed to predict the effects of drug combinations, their dosages, their planning. This model could also be useful in several types of intervention such as the invalidation or deletion of a gene. The model, called Compositional Perturbation Autoencoder (CPA), is licensed as an open-source tool and includes an API and a Python package.
Research to perfect drug combinations
In order to treat malignant tumors of cancer patients, a combination of drugs is prepared beforehand. The CPA is the solution proposed by Facebook AI and the Helmholtz Zentrum München to find an effective combination of existing drugs with an appropriate dosage. This is a major problem when you consider that the possibilities can range from 5,000 to 19 billion combinations.
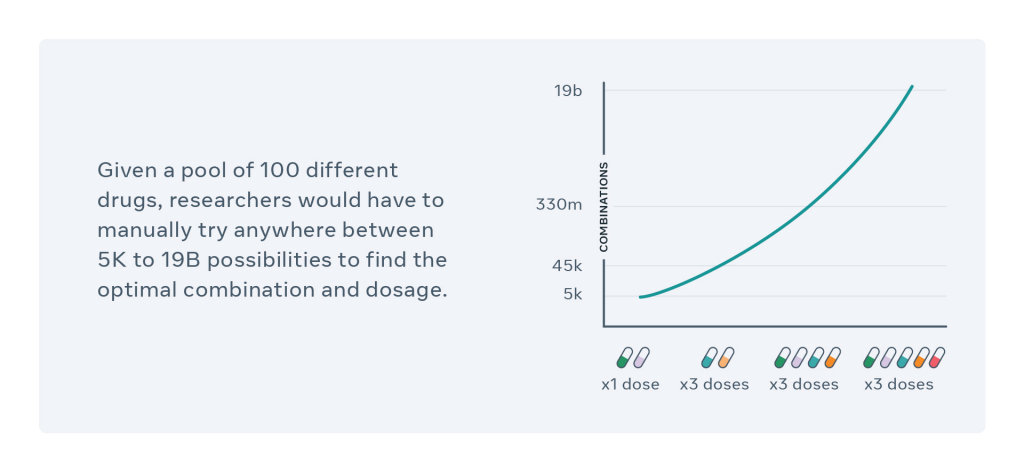
This problem was the starting point of a long work, all the details of which are proposed in a publication published on April 14. This work is the logical extension of those already carried out on computational methods related to drug interactions included in the learning data set.
This research was conducted by Mohammad Lotfollahi, Anna Klimovskaia Susmelj, Carlo De Donno, Yuge Ji, Ignacio L. Ibarra, F. Alexander Wolf, Nafissa Yakubova, Fabian J. Theis and David Lopez-Paz, all of whom are authors of the above-mentioned paper.
The solution: self-supervised learning
Self-supervised learning is the solution that Facebook AI and the Helmholtz Zentrum München have been working on in order to narrow down the range of possible combinations of correctly dosed drugs.
One issue that several researchers and scientists have raised concerns single-cell RNA sequencing. Single cell RNA sequencing is used as a method of measuring RNA gene expression in individual cells at the molecular level. With this, it is possible to study the effects of different perturbations such as drug combinations or gene deletion.
Until now, there has been no effective approach to predict the effects of novel drug combinations and other perturbations. With single-cell RNA sequencing, researchers have developed and published datasets using this technique to advance biomedical research.
But the whole point of this data is that it should be analyzed to get the most benefit from it and to limit the possible combinations. Machine learning models can be a solution to do this kind of analysis, but they need to be able to generalize and extrapolate interesting aspects of a data structure without relying on characterized training data to make predictions about new conditions.
The solution developed by Facebook AI and the Helmholtz Zentrum München is to use self-encoding. The computer synthesizes the data to build useful forecasting models. This self-supervised learning technique aims to compress and decompress the data, all based on the uncharacterized gene expression vectors of different pathologies
The system relies on the isolation of key features of a cell such as the effects of a drug on it, a particular dosage, a schedule, a combination, a gene deletion or a specific cell type. The autoencoding model autonomously recombines these features to predict their effects on the cell’s gene expressions.
The model is trained such that the output of the decoder matches the gene expression initially available in the single-cell RNA-seq data set. There are then three steps characteristic to the learning of this model:
- The encoder network transforms the gene expression associated with a cell in the data set into a “cell representation”. An integration network translates the processing applied to that cell into a “processing representation”.
- By combining these cell and processing representations, a bottleneck representation is realized.
- A decoder network translates the bottleneck representation into a gene expression vector.
CPA: a self-supervision technique
The CPA uses a self-supervision technique to observe cells treated (e.g. tumor cells) with a limited number of drug combinations and predicts the effects of novel combinations. Let’s say we have four types of cells that we’ll call A, B, C, and A+B, and we have data that tells us how the drugs react on each of these cells. With the CPA, doctors can deduce the impact that each drug has on each cell type and will even recombine that data to extrapolate A+C and B+C combinations or even interactions between A+B and C+D.
<img class=”alignnone size-full wp-image-27011″ src=”https://www.actuia.com/wp-content/uploads/2021/04/Schema-2-Extrapolation-de-combinaisons-et-interractions-entre-les-cellules-selon-le-medicament-utilise.jpg” alt=”The CPA will be able to tell the impact of drugs on one or more cell types that can interact with each other or even be extrapolated. width=”1024″ height=”499″ /> “CPA can generate hypotheses, which can be used to
The CPA can generate hypotheses, guide experimental design processes for new drugs, and target interesting combinations to reduce the millions or even billions of possible combinations, thus avoiding the need for researchers to test all of these combinations. The CPA therefore represents a significant time saving in addition to offering concrete expertise.
This self-encoding model was applied to five RNA-seq datasets containing measurements and results for different drugs, assays and other perturbations observed on cancer cells. Its performance was measured using an R2 indicator representing the quality of predictions regarding the expression of individual genes.
The results are as follows: the R2 scores remained constant for the training and test sets and increased for the non-distribution sets. This means that the CPA predictions of the effects of key drug combinations and assays on cancer cells reliably match those obtained in the test data set.
To test the limitations of the model, the amount of training data was reduced and the amount of non-distribution data was increased. Despite a considerable decrease in performance, the predictions were far from random.
Translated from Facebook AI et le HZM utilisent l’intelligence artificielle pour prévoir les effets des combinaisons de médicaments









